Syllabus (Fourth Edition, 2023)
Topics
T1: General Microbiology
i. Describe the classification of bacterium.
ii. Describe the principles of anti-microbial resistance.
iii. Broadly outline the classification of viruses and fungi.
T2: Antimicrobial Pharmacology
i. Describe the classification and pharmacology of antibacterial agents.
ii. Describe the classification and pharmacology of antiviral and antifungal agents.
iii. Outline the pharmacology of antiseptics and disinfectants.
Topics not covered in previous SAQs
T1: General Microbiology
iii. Broadly outline the classification of viruses and fungi.
T2: Antimicrobial Pharmacology
ii. Describe the classification and pharmacology of antiviral and antifungal agents.
Learning Objectives for the First Part Examination in Intensive Care Medicine
- This will ensure that trainees, tutors, and examiners can work from a common base.
- All examination questions are based around this Syllabus.
- These learning objectives are designed to outline the minimum level of understanding required for each topic.
- The accompanying texts are recommended on the basis that the material contained within them provides sufficient information for trainees to meet the learning objectives.
- Trainees are strongly encouraged to explore the existing and evolving body of knowledge of the Basic Sciences as they apply to Intensive Care Medicine by reading widely.
- For all sections of the syllabus an understanding of normal physiology and physiology at extremes of age, obesity, pregnancy (including foetal) and disease (particularly critical illness) is expected.
- Similarly, for pharmacology, trainees are expected to understand a drug’s pharmacology in these contexts.
- An understanding of potential toxicity and relevant antidotes is also expected.
Definitions
Throughout the document specific wording has been used under the required abilities to indicate the level of knowledge and understanding expected and a glossary of these terms is provided.
Definitions
| Calculate | Work out or estimate using mathematical principles. |
| Classify | Divide into categories; organise, arrange. |
| Compare and contrast | Examine similarities and differences. |
| Define | Give the precise meaning. |
| Describe | Give a detailed account of. |
| Explain | Make plain. |
| Interpret | Explain the meaning or significance. |
| Outline | Provide a summary of the important points. |
| Relate | Show a connection between. |
| Understand | Appreciate the details of; comprehend. |
SAQs
T1: General Microbiology
i. Describe the classification of bacterium.
2014A 10
Classify bacteria according to the Gram stain system and their shape.
Give two examples for each classification. (40% of marks)
List with examples the mechanisms of bacterial antibiotic resistance. (60% of marks)
2008B 16
Classify bacteria according to the Gram stain system and the shape of the bacteria, and give two examples for each classification (40% of marks).
Outline the different mechanisms of bacterial antibiotic resistance and an antibiotic for which that mechanism may apply (60% of marks).
CICMWrecks Answer: Classify Bacteria
Gram Stain
Method of differentiated bacteria according to their cell wall characteristics, using
staining with crystal violet dye
- Gram positive bacteria have a thick outer cell wall composed of peptidoglycan,
which stains positive with crystal violet dye - Gram negative bacteria have an outer cell membrane enclosing a thinner
peptidoglycan cell wall, which has decreased affinity to the crystal violet dye
Classification of bacteria
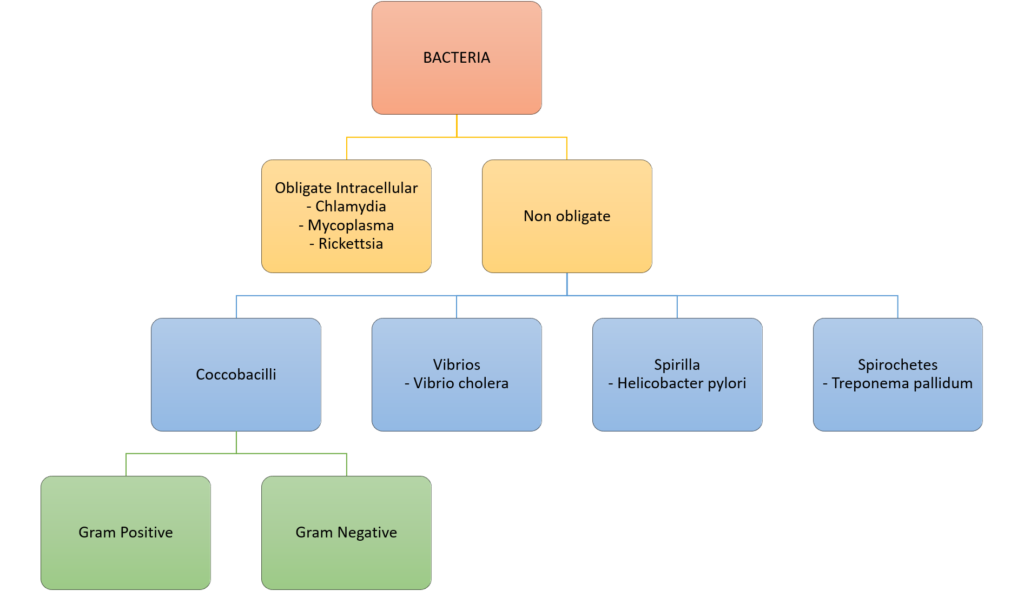
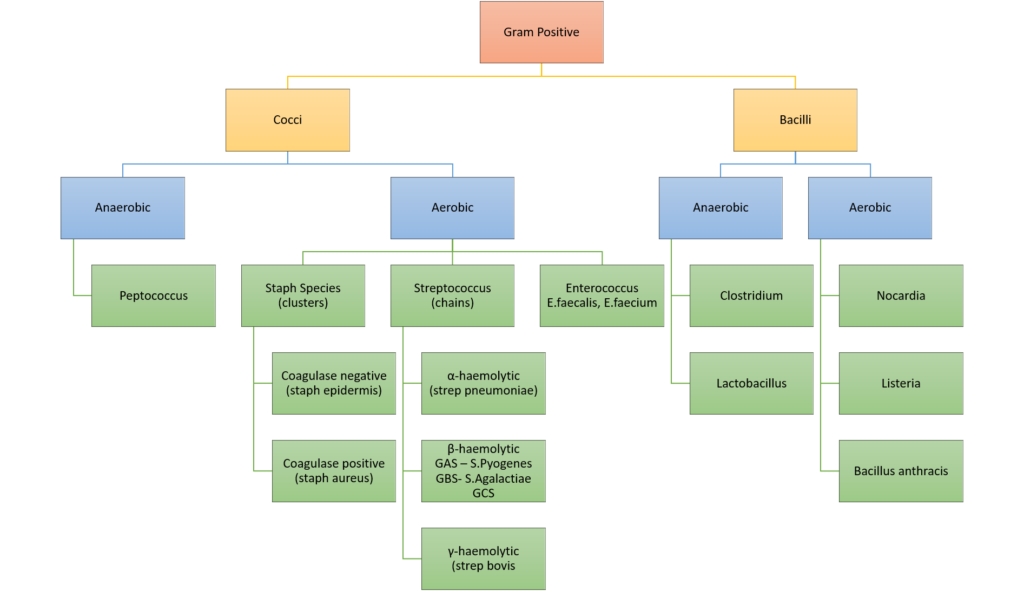
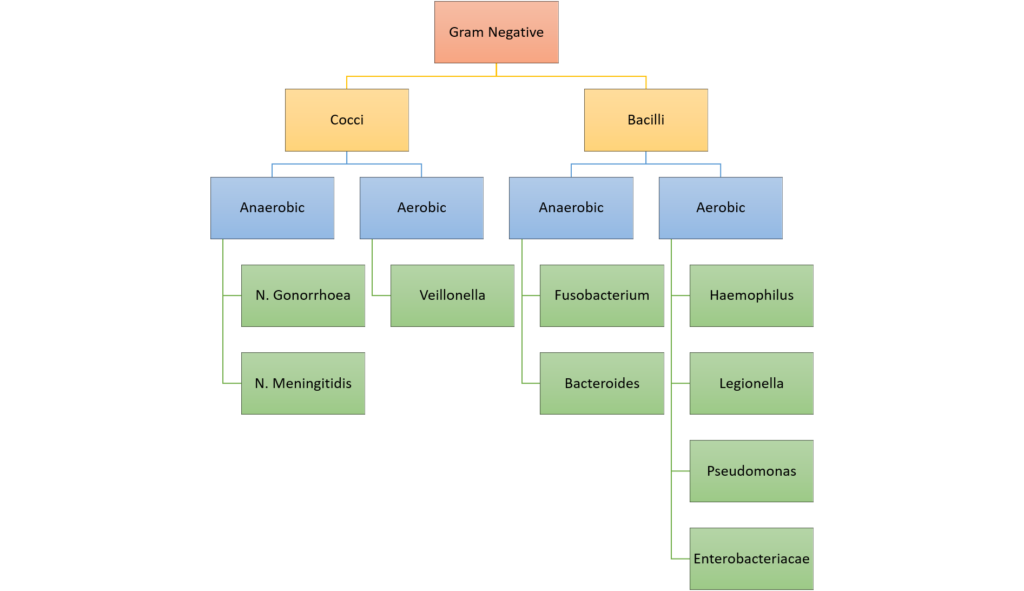
Gladwin / Sakurai / JC 2020
CICMWrecks Answer: Antibiotic resistance
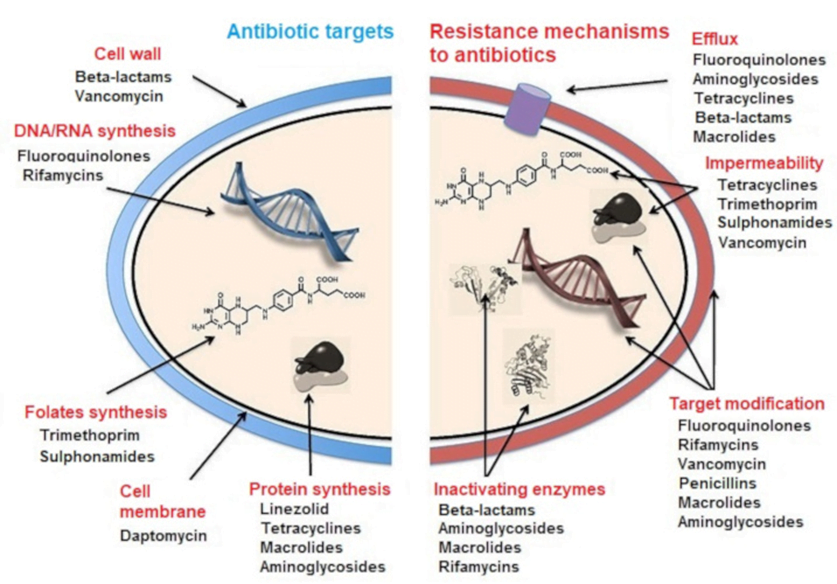
MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2014A 10: 63% of candidates passed this question.
Generally candidates provided an accurate classification of bacteria based upon Gram staining and shape. None or poorly staining organisms were often overlooked. Mechanism of resistance was also well covered, with examples. Areas of weakness were a lack of descriptive detail and/or omission of an example for each mechanism. Mechanisms such as metabolic blockade of essential pathways for antibacterial action and active removal of intracellular antibiotic were more commonly omitted.
2008B 16: 3 (60%) candidates passed this question
This question also highlighted the importance of candidates noting the way marks were proportioned. A good answer required the following points – Classification and examples – gram-positive cocci (Staphylococcus aureus, Streptococcus pyogenes or pneumoniae or agalactiae), gram-negative cocci (Neisseria meningitidis, N. gonorrhoeae), gram-positive bacilli (Bacillus anthracis, Listeria, corynebacteria, Clostridium difficile, etc), gram-negative bacilli (Escherichia Coli or E. Coli, Proteus, Yersinia, Salmonella, Shigella, Pseudomonas aeruginosa, Klebsiella pneumoniae,
Legionella).
Mechanisms of resistance include: a) Enzyme inactivation, beta-lactamase or Extended Spectrum Beta-lactamase b) Enzyme addition: enzyme produced by bacteria that add a chemical group to the antibiotic to inhibit its activity, aminoglycoside resistance by Staphylococcus aureus or Pseudomonas. c) Impermeability, anaerobes have no oxygen dependent transport mechanism which stops the penetration of aminoglycosides into the bacteria. d) Efflux mechanisms by acquisition of an inner member protein which actively pumps antibiotics out of the cell, E.Coli becomes resistance to tetracycline by this mechanism. e) Alternative pathway to circumvent the metabolic block impose by antibiotic, Some Staphylococcus aureus are resistant to methicillin by developing or acquiring the gene mecA which produces an alternative penicillin binding protein and hence they are not
inhibited by methicillin. f) Alteration of the target site, rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene.
Syllabus: M2a, b, c
Reference Text: Microbiology and Infection at a Glance by Gillespie & Bamford 3rd Ed, 2007 page 8-9,21.
2024B 07
(a) Categorise bacteria by their gram stain appearance and shape. Give TWO examples of bacteria for each group (30% of marks).
(b) Provide TWO examples of antimicrobials used against each group and outline their mechanism of action (20% of marks).
(c) Outline the mechanisms of bacterial resistance with ONE example for each (50% of marks).
CICMWrecks Answer: Classify Bacteria and antimicrobials
Gram Stain
Method of differentiated bacteria according to their cell wall characteristics, using
staining with crystal violet dye
- Gram positive bacteria have a thick outer cell wall composed of peptidoglycan,
which stains positive with crystal violet dye - Gram negative bacteria have an outer cell membrane enclosing a thinner
peptidoglycan cell wall, which has decreased affinity to the crystal violet dye
Classification of bacteria



Examples of Antimicrobials + MoA
| Group | Antimicrobials | Mechanism of Action |
|---|---|---|
| Gram Positive Cocci | Penicillin | β lactam antibiotic Binds to penicillin binding protein (transpeptidase) and prevents crosslinking of bacterial peptidoglycan impairs cell wall synthesis |
| Vancomycin | Inhibits Glycopeptide synthetase prevents peptidoglycan formation in bacterial cell well (Unlike penicillins, prevents the transfer and addition of the muramylpentapeptide building blocks that make up the peptidoglycan molecule itself.) May also alter membrane permeability and selectively inhibit RNA synthesis. | |
| Gram Positive Bacilli | Clindamycin | Inhibitor of bacterial 50S ribosomal subunit Prevents protein synthesis |
| Erythromycin | Inhibits 50s subunit to prevent protein synthesis Bacteriocidal/static | |
| Gram Negative Cocci | Ceftriaxone | Beta lactam ring binds to multiple penicillin binding proteins (carboxy/endo/ transpeptidase) and prevents crosslinking of bacterial peptidoglycan inhibits cell wall synthesis BROADER SPECTRUM Bacteria eventually lyse due to ongoing activity of cell wall autolytic enzymes (autolysins and murein hydrolases) while cell wall assembly is arrested Configuration: Stability against Betalactamases |
| Ciprofloxacin | Bacteriocidal antimicrobials that block DNA replication by blocking tropoisomerase enzymes, which are essential for the supercoiling, replication and separation of circular bacterial DNA | |
| Gram Negative Bacilli | Gentamicin | bactericidal Binds to the bacterial 30S ribosomal subunit to inhibit protein synthesis and thus bacterial growth |
| Meropenem | Binds to several penicillin binding protein and prevents crosslinking of bacterial peptidoglycan inhibits cell wall synthesis BROADEST SPECTRUM Bacteria eventually lyse due to ongoing activity of cell wall autolytic enzymes (autolysins and murein hydrolases) while cell wall assembly is arrested Configuration: Stability against Betalactamases and ESBLs |
Gladwin / Sakurai / JC 2020
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2024B 07: 49% of candidates passed this question.
The first part of the question asked candidates to categorise bacteria by gram stain and shape. A description of the gram stain procedure or discussion of bacterial cell wall characteristics was not required. Grouping of bacteria into gram positive cocci, gram positive bacilli, gram negative cocci, gram negative bacilli and others was expected with appropriate examples. A list of two antimicrobials that can be used against each of these classifications followed and a brief outline of their mechanism of action was expected. The largest weighting of marks was reserved for the third component of the question. Mechanisms of resistance included innate and acquired. Aspects of acquired resistance expected were genetic factors that alter the bacterial target, changes in bacterial permeability/efflux pumps and presence of enzymes that modify or negate the drug.
2015A 05
Classify gram positive bacteria with examples. (20% of marks)
Outline the pharmacology of vancomycin (80% of marks).
CICMWrecks Answer: Gram positive Bacteria
GRAM POSITIVE BACTERIA
Gram Stain
Method of differentiated bacteria according to their cell wall characteristics, using
staining with crystal violet dye
- Gram positive bacteria have a thick outer cell wall composed of peptidoglycan,
which stains positive with crystal violet dye - Gram negative bacteria have an outer cell membrane enclosing a thinner
peptidoglycan cell wall, which has decreased affinity to the crystal violet dye

Gladwin / Sakurai / JC 2020
CICMWrecks Answer: Pharmacology: Vancomycin
PHARMACOLOGY OF VANCOMYCIN
Examiner Comments
2015A 05: 75 % of candidates passed this question.
The classification should have demonstrated a framework that covered relevant gram positive pathogens. Examples should have included both genus and species. More detail than “strep” or “Staph” was expected.
Knowledge of vancomycin was expected to include an outline of pharmaceutics, pharmacodynamics, pharmacokinetics, dosing and adverse effects. In particular, pharmacokinetics should be well understood as there are significant implications for dosing.
Common errors included incorrect examples such as “clostridium” or classifications lacking detail with respect to examples.
ii. Describe the principles of anti-microbial resistance.
2014A 10
Classify bacteria according to the Gram stain system and their shape. Give two examples for each classification. (40% of marks)
List with examples the mechanisms of bacterial antibiotic resistance. (60% of marks)
2008B 16
Classify bacteria according to the Gram stain system and the shape of the bacteria, and give two examples for each classification (40% of marks).
Outline the different mechanisms of bacterial antibiotic resistance and an antibiotic for which that mechanism may apply (60% of marks).
CICMWrecks Answer: Classify Bacteria
Gram Stain
Method of differentiated bacteria according to their cell wall characteristics, using
staining with crystal violet dye
- Gram positive bacteria have a thick outer cell wall composed of peptidoglycan,
which stains positive with crystal violet dye - Gram negative bacteria have an outer cell membrane enclosing a thinner
peptidoglycan cell wall, which has decreased affinity to the crystal violet dye
Classification of bacteria



Gladwin / Sakurai / JC 2020
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2014A 10: 63% of candidates passed this question.
Generally candidates provided an accurate classification of bacteria based upon Gram staining and shape. None or poorly staining organisms were often overlooked. Mechanism of resistance was also well covered, with examples. Areas of weakness were a lack of descriptive detail and/or omission of an example for each mechanism. Mechanisms such as metabolic blockade of essential pathways for antibacterial action and active removal of intracellular antibiotic were more commonly omitted.
2008B 16: 3 (60%) candidates passed this question
This question also highlighted the importance of candidates noting the way marks were proportioned. A good answer required the following points – Classification and examples – gram-positive cocci (Staphylococcus aureus, Streptococcus pyogenes or pneumoniae or agalactiae), gram-negative cocci (Neisseria meningitidis, N. gonorrhoeae), gram-positive bacilli (Bacillus anthracis, Listeria, corynebacteria, Clostridium difficile, etc), gram-negative bacilli (Escherichia Coli or E. Coli, Proteus, Yersinia, Salmonella, Shigella, Pseudomonas aeruginosa, Klebsiella pneumoniae,
Legionella).
Mechanisms of resistance include: a) Enzyme inactivation, beta-lactamase or Extended Spectrum Beta-lactamase b) Enzyme addition: enzyme produced by bacteria that add a chemical group to the antibiotic to inhibit its activity, aminoglycoside resistance by Staphylococcus aureus or Pseudomonas. c) Impermeability, anaerobes have no oxygen dependent transport mechanism which stops the penetration of aminoglycosides into the bacteria. d) Efflux mechanisms by acquisition of an inner member protein which actively pumps antibiotics out of the cell, E.Coli becomes resistance to tetracycline by this mechanism. e) Alternative pathway to circumvent the metabolic block impose by antibiotic, Some Staphylococcus aureus are resistant to methicillin by developing or acquiring the gene mecA which produces an alternative penicillin binding protein and hence they are not
inhibited by methicillin. f) Alteration of the target site, rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene.
Syllabus: M2a, b, c
Reference Text: Microbiology and Infection at a Glance by Gillespie & Bamford 3rd Ed, 2007 page 8-9,21.
2024B 07
(a) Categorise bacteria by their gram stain appearance and shape. Give TWO examples of bacteria for each group (30% of marks).
(b) Provide TWO examples of antimicrobials used against each group and outline their mechanism of action (20% of marks).
(c) Outline the mechanisms of bacterial resistance with ONE example for each (50% of marks).
CICMWrecks Answer: Classify Bacteria and antimicrobials
Gram Stain
Method of differentiated bacteria according to their cell wall characteristics, using
staining with crystal violet dye
- Gram positive bacteria have a thick outer cell wall composed of peptidoglycan,
which stains positive with crystal violet dye - Gram negative bacteria have an outer cell membrane enclosing a thinner
peptidoglycan cell wall, which has decreased affinity to the crystal violet dye
Classification of bacteria



Examples of Antimicrobials + MoA
| Group | Antimicrobials | Mechanism of Action |
|---|---|---|
| Gram Positive Cocci | Penicillin | β lactam antibiotic Binds to penicillin binding protein (transpeptidase) and prevents crosslinking of bacterial peptidoglycan impairs cell wall synthesis |
| Vancomycin | Inhibits Glycopeptide synthetase prevents peptidoglycan formation in bacterial cell well (Unlike penicillins, prevents the transfer and addition of the muramylpentapeptide building blocks that make up the peptidoglycan molecule itself.) May also alter membrane permeability and selectively inhibit RNA synthesis. | |
| Gram Positive Bacilli | Clindamycin | Inhibitor of bacterial 50S ribosomal subunit Prevents protein synthesis |
| Erythromycin | Inhibits 50s subunit to prevent protein synthesis Bacteriocidal/static | |
| Gram Negative Cocci | Ceftriaxone | Beta lactam ring binds to multiple penicillin binding proteins (carboxy/endo/ transpeptidase) and prevents crosslinking of bacterial peptidoglycan inhibits cell wall synthesis BROADER SPECTRUM Bacteria eventually lyse due to ongoing activity of cell wall autolytic enzymes (autolysins and murein hydrolases) while cell wall assembly is arrested Configuration: Stability against Betalactamases |
| Ciprofloxacin | Bacteriocidal antimicrobials that block DNA replication by blocking tropoisomerase enzymes, which are essential for the supercoiling, replication and separation of circular bacterial DNA | |
| Gram Negative Bacilli | Gentamicin | bactericidal Binds to the bacterial 30S ribosomal subunit to inhibit protein synthesis and thus bacterial growth |
| Meropenem | Binds to several penicillin binding protein and prevents crosslinking of bacterial peptidoglycan inhibits cell wall synthesis BROADEST SPECTRUM Bacteria eventually lyse due to ongoing activity of cell wall autolytic enzymes (autolysins and murein hydrolases) while cell wall assembly is arrested Configuration: Stability against Betalactamases and ESBLs |
Gladwin / Sakurai / JC 2020
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2024B 07: 49% of candidates passed this question.
The first part of the question asked candidates to categorise bacteria by gram stain and shape. A description of the gram stain procedure or discussion of bacterial cell wall characteristics was not required. Grouping of bacteria into gram positive cocci, gram positive bacilli, gram negative cocci, gram negative bacilli and others was expected with appropriate examples. A list of two antimicrobials that can be used against each of these classifications followed and a brief outline of their mechanism of action was expected. The largest weighting of marks was reserved for the third component of the question. Mechanisms of resistance included innate and acquired. Aspects of acquired resistance expected were genetic factors that alter the bacterial target, changes in bacterial permeability/efflux pumps and presence of enzymes that modify or negate the drug.
2009B 01
Explain the mechanisms by which a bacterium may become resistant to an antimicrobial agent and provide an example (organism and antimicrobial) of each mechanism. (60% of marks)
How is antimicrobial resistance spread? (40% of marks)
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2009B 01: 6 (67%) of candidates passed this question.
Candidates were expected to explain and provide an example of an antimicrobial agent for mechanisms such as enzyme inactivation of the antimicrobial, alteration of antimicrobial binding/target sites, reduction of antimicrobial drug uptake or active efflux of drug by the bacteria and alteration in enzymatic pathways. Use of a table aided candidates expressing this information. Examples of the methods of spread of antibiotic resistance in bacteria that were expected were: transfer of resistant bacteria from person to person; horizontal gene transfer; transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria); transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells; conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Most candidates could name various mechanisms of resistance but many failed to gain marks as they did not provide an example of an organism and antimicrobial. The second part of the question concerning the spread of resistance was generally done quite well.
Syllabus – M2a2c
Reference: Pharmacology, Rang and Dale Ch 46, Goodman & Gillman, Ch42
2020A 05
Outline the mechanisms of antimicrobial resistance (50% of marks).
Briefly outline the pharmacology of ciprofloxacin (50% of marks).
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
CICMWrecks Answer: Pharmacology: Ciprofloxacin
PHARMACOLOGY OF CIPROFLOXACIN
Examiner Comments
2020A 05: 71% of candidates passed this question.
Most candidates had a structured answer to mechanisms of resistance that covered the major categories (alter target protein, prevent entry, efflux, degrade drug) and provided an example of a bacteria and the affected antibiotic, as was required to answer the question in full. Ciprofloxacin, whilst perhaps not a first line drug in the ICU, was not well known by many candidates. Better answers included a brief outline of class, mechanism of action (action on DNA gyrase to inhibit replication), spectrum (Gram negatives particularly mentioning Pseudomonas, lesser Grampositive cover, not anaerobes, some atypical), PK (with correct dose, wide penetration into tissues including bone/prostate etc., predominantly renal excretion), side effects/toxicity (common or specific to cipro e.g. QT, tendinitis, arthropathy) and an example of resistance.
2019A 09
Classify antibiotics with respect to their mechanism of action (50% of marks).
Outline the mechanisms of antimicrobial resistance (50% of marks). Give specific examples of each.
CICMWrecks Answer: Antibiotic Classification by MoA

CLASSIFICATION OF ANTIBIOTICS
| 1. Inhibitor of cell wall synthesis/ Peptidoglycan Inhibitors: | · Beta-lactams: Penicillin · Cephalosporins: Ceftriaxone · Carbapenems: Meropenem · Glycopeptides: Vancomycin |
| 2. Inhibitor of Nucleic acid synthesis: | · Quinolones: Ciprofloxacin · Rifamycins: Rifampicin · Nitroimidazoles: Metronidazole · Nitrofurantoin |
| 3. Inhibitor of folic acid synthesis (Folate antagonistic) | · Sulfonamides: Sulfamethoxazole · Aminopyrimidines: Trimethoprim |
| 4. Inhibitor of cytoplasmic membrane: | · Lipopeptide: Daptomycin · Polymyxins: Colistin |
| 5. Inhibitor of protein synthesis: | · Aminoglycosides: Gentamicin · Lincosamides: Clindamycin · Macrolides: Erythromycin · Tetracyclines: Tetracycline |
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2019A 09: 70% of candidates passed this question.
This question was well answered. Marks were awarded for correct pairing of mechanism of action and resistance with examples of drug class. Few mentioned the mechanisms by which resistance is present; acquired or generated.
iii. Broadly outline the classification of viruses and fungi.
2010A 12
Outline the classification of viruses giving examples of each class (60% marks). Describe the mechanism of action of acyclovir and oseltamivir (40% marks).
CICMWrecks Answer
Baltimore Classification system (for Viruses)
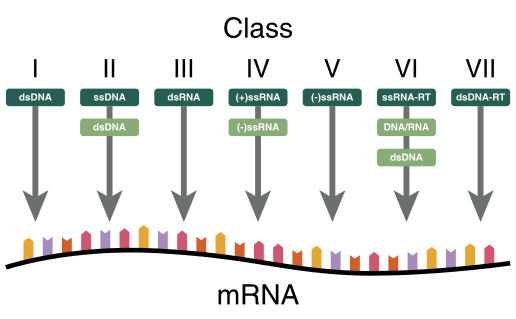
| I | dsDNA viruses | mRNA is transcribed directly from the DNA template | Adenoviruses, Herpesviruses, Poxviruses |
| II | ssDNA viruses | (+ strand or “sense”) DNA DNA is converted to double stranded form before RNA is transcribed | Parvovirus (B19) |
| III | dsRNA viruses | mRNA is transcribed from the RNA genome | Rotavirus |
| IV | (+)ssRNA viruses | (+ strand or sense) RNA Genome functions as mRNA | Picornaviruses (Entero, HepA), Flaviviruses (Dengue) |
| V | (−)ssRNA viruses | (− strand or antisense) RNA mRNA is transcribed from the RNA genome | Orthomyxoviruses (Influenza) |
| VI | ssRNA-RT viruses | (+ strand or sense) RNA with DNA intermediate in life-cycle RT makes DNA from the RNA genome DNA is then incorporated into host genome mRNA is transcribed from the incorporated DNA | Retroviruses (HIV) |
| VII | dsDNA-RT viruses | DNA with RNA intermediate in life-cycle Viral DNA is replicated through an RNA intermediate, the RNA may serve directly as mRNA or as a template to make mRNA | Hep B |
Key:
ss: Single stranded
ds: Double stranded
(-): – strand or antisense
(+): + strand or sense
RT – Reverse Transcriptase
Past Classifications (now not used):
By shape:
- filamentous
- isometric (or icosahedral)
- envelopedH
- Head and tail
By Genome Structure and Core:
- RNA / DNA
- Single-stranded / Double stranded
- Linear/Circular
- Non-segmented/Segmented
By capsid structure:
- Naked icosahedral – Hep A, Polio
- Enveloped icosahedral – EBV, HSV
- Enveloped helical – Influenza, Measles
- Naked Helical – Tobacco Mosaic virus
- Complex – Herper, Smallpox, Hep B
Mechanism of Action: Oseltamivir vs Aciclovir
| Oseltamivir | Aciclovir |
|---|---|
| Neuraminidase inhibitor (Anti-Influenza) | Viral guanosine analogue |
| Prodrug | Prodrug |
| Active metabolite inhibits viral Neuraminidase | Converted to aciclovir monophosphate by viral thymidine kinase which in turn is converted to aciclovir triphosphate by host cell |
| cleaves the sialic acid (which is found on glycoproteins on the surface of human cells that helps new virions to exit the cell) | ACV-TP is HSV-specified DNA polymerase inhibitor |
| prevents viral release by infected cells (viral shedding) | inhibits nucleic acid production, incorporates into DNA resulting in chain termination |
| Active against Influenza | Active against herpesvirus family: HSV-1,2, VZV,EBV,CMV(least) |
Mooney / JC 2020
Examiner Comments
2010A 12: 1 (10%) of candidates passed this question
Viruses are classified according to:
a) genetic material
b) mode replication
c) structural proteins (capsids)
d) presence of an envelope
Thus DNA viruses, double or single stranded DNA usually replicate in the nucleus of the host cell via polymerase, not incorporated into the host genetic material. Examples being double stranded, herpes, adenovirus, poxvirus and single stranded, parvovirus. In comparison, RNA viruses, single strand and have 2 different reproduction strategies, being RNA sense(positive) and RNA antisense(negative), an example is paramyxovirus. For retroviruses, the single stranded RNA can’t act as mRNA and is transcribed into DNA by a reverse transcriptase. This DNA is incorporated into the host DNA, so the host makes the viral RNA, for example HIV.
Candidates were also expected to briefly mention capsids and viral envelopes. In relation to the second part of the question, candidates were expected to mention that acyclovir inhibits DNA polymerase in the terminal nucleic acid chain and that oseltamivir is a neuraminidase inhibitor which prevents the budding of new viruses from the infected cells. Most candidates had very little knowledge of this area.
Syllabus: M2 2a&d 8 Reference: Medical Microbiology and Infection at a Glance, Gillespie &, Bamford pgs 58,59, Basic and Clinical Pharmacology, Katzung pgs791,815
T2: Antimicrobial Pharmacology
i. Describe the classification and pharmacology of antibacterial agents.
Classifications
2019A 09
Classify antibiotics with respect to their mechanism of action (50% of marks).
Outline the mechanisms of antimicrobial resistance (50% of marks). Give specific examples of each.
CICMWrecks Answer: Antibiotic Classification by MoA

CLASSIFICATION OF ANTIBIOTICS
| 1. Inhibitor of cell wall synthesis/ Peptidoglycan Inhibitors: | · Beta-lactams: Penicillin · Cephalosporins: Ceftriaxone · Carbapenems: Meropenem · Glycopeptides: Vancomycin |
| 2. Inhibitor of Nucleic acid synthesis: | · Quinolones: Ciprofloxacin · Rifamycins: Rifampicin · Nitroimidazoles: Metronidazole · Nitrofurantoin |
| 3. Inhibitor of folic acid synthesis (Folate antagonistic) | · Sulfonamides: Sulfamethoxazole · Aminopyrimidines: Trimethoprim |
| 4. Inhibitor of cytoplasmic membrane: | · Lipopeptide: Daptomycin · Polymyxins: Colistin |
| 5. Inhibitor of protein synthesis: | · Aminoglycosides: Gentamicin · Lincosamides: Clindamycin · Macrolides: Erythromycin · Tetracyclines: Tetracycline |
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
Examiner Comments
2019A 09: 70% of candidates passed this question.
This question was well answered. Marks were awarded for correct pairing of mechanism of action and resistance with examples of drug class. Few mentioned the mechanisms by which resistance is present; acquired or generated.
Anti-Staph / Gram Positive
2022B 09
List any five classes of antibiotics with anti-staphylococcal activity and provide one example of each class. Outline the mechanism of action and side effects of the five drugs.
CICMWrecks Answer
Anti-staphylococcal antibiotics
| Class | Antibiotic | Mechanism of Action | Side Effects |
|---|---|---|---|
| Penicillin | Flucloxacillin | Beta lactam ring binds to penicillin binding protein (transpeptidase) and prevents crosslinking of bacterial peptidoglycan inhibits cell wall synthesis Bacteria eventually lyse due to ongoing activity of cell wall autolytic enzymes (autolysins and murein hydrolases) while cell wall assembly is arrested Less bactericidal but stable to staph beta-lactamases | Up to 10% of the population have allergies to penicillins. Due to the high percentage excreted renally unchanged dose adjustment is required in low urine output states. Severe cholestatic hepatitis has been reported idiosyncratically. |
| Glycopeptide | Vancomycin | Inhibits Glycopeptide synthetase prevents peptidoglycan formation in bacterial cell well (Unlike penicillins, prevents the transfer and addition of the muramylpentapeptide building blocks that make up the peptidoglycan molecule itself.) May also alter membrane permeability and selectively inhibit RNA synthesis. Antimicrobial activity Dependent on Duration above MIC, not concentration | Hypersensitivity reactions including anaphylaxis. Rapid infusion is associated with histamine release – red-man syndrome. Ototoxicity rare, dose related. Nephrotoxicity has declined with improved formulations, usually resolves with cessation of drug |
| Cephalosporin | Cefazolin | Beta lactam ring binds to multiple penicillin binding proteins (carboxy/endo/ transpeptidase) and prevents crosslinking of bacterial peptidoglycan inhibits cell wall synthesis BROADER SPECTRUM Bacteria eventually lyse due to ongoing activity of cell wall autolytic enzymes (autolysins and murein hydrolases) while cell wall assembly is arrested Configuration: Stability against Betalactamases | Generally well tolerated Hypersensitivity reactions (10% cross reactivity with penicillin allergies), thrombophlebitis, interstitial nephritis, low prothrombin, flushing and headaches with EtOH. |
| Lincosamide | Clindamycin | Inhibitor of bacterial 50S ribosomal subunit Prevents protein synthesis | – Pseudomembranous colitis due to c.diff – Contact dermatitis – Metallic taste – Anaphylaxis |
| Linezolid | Oxazolidinone | inhibits bacterial protein synthesis by binding specifically to the 50s ribosomal subunit, thereby preventing initiation complex formation. | Headache, LFT derangement, Taste Alteration, GI disturbances Skin/bleeding problems, phlebitis, pancreatitis |
Daptomycin, Rifampicin
Examiner Comments
2022B 09: 82% of candidates passed this question.
In general, the question was well answered. Better answers provided examples of antibiotics from different categories of mechanism of action and were able to describe specific side effects relevant or unique to that antibiotic. Mechanisms of action that were generic in terms of site of action attracted fewer marks. Some examples of antibiotics that candidates provided were not anti-staphylococcal antibiotics (eg; Benzyl Penicillin, Clavulanic acid). The examiners commented that the use of a tabular format as an answer template resulted in answers scoring marks in most time efficient manner. Frequent omissions or commissions resulting in lost marks noted by the examiners were as follows; many papers showed a lack of detail regarding the precise mechanism of action of penicillins particularly around inhibition of transpeptidases whilst the use of non-specific side effects such as nausea vomiting and diarrhoea did not attract marks.
2015A 05
Classify gram positive bacteria with examples. (20% of marks)
Outline the pharmacology of vancomycin (80% of marks).
CICMWrecks Answer: Gram positive Bacteria
GRAM POSITIVE BACTERIA
Gram Stain
Method of differentiated bacteria according to their cell wall characteristics, using
staining with crystal violet dye
- Gram positive bacteria have a thick outer cell wall composed of peptidoglycan,
which stains positive with crystal violet dye - Gram negative bacteria have an outer cell membrane enclosing a thinner
peptidoglycan cell wall, which has decreased affinity to the crystal violet dye

Gladwin / Sakurai / JC 2020
CICMWrecks Answer: Pharmacology: Vancomycin
PHARMACOLOGY OF VANCOMYCIN
Examiner Comments
2015A 05: 75 % of candidates passed this question.
The classification should have demonstrated a framework that covered relevant gram positive pathogens. Examples should have included both genus and species. More detail than “strep” or “Staph” was expected.
Knowledge of vancomycin was expected to include an outline of pharmaceutics, pharmacodynamics, pharmacokinetics, dosing and adverse effects. In particular, pharmacokinetics should be well understood as there are significant implications for dosing.
Common errors included incorrect examples such as “clostridium” or classifications lacking detail with respect to examples.
2013B 16
Describe the pharmacology of vancomycin.
2012A 12
Outline the pharmacology of vancomycin
Examiner Comments
2013B 16: 16 candidates passed (59.3%).
A commonly used drug in intensive care practice, for which a high level of understanding is required (Level A). In general answers were sufficient for a pass, but there was still a lack of sufficient breadth of knowledge, in particular to pharmacokinetics and detailed mechanism of action.
2012A 12: 6 (60%) of candidates passed.
A basic and fundamental pharmacology question which required candidates to present their answer in a coherent fashion, as well as demonstrate sufficient knowledge. Candidates were expected to mention spectrum and mechanism of action, pharmacokinetics (including dose, distribution, elimination, etc) and adverse effects, activity profile e.g. time-dependent, antimicrobial activity depends on the duration that the serum drug concentration exceeds the minimum inhibitory concentration (MIC) of the target organism and not concentration dependence.
2018B 10
Compare and contrast the pharmacology of vancomycin and flucloxacillin.
2016A 22
Compare and contrast the mechanism of action (25% of marks), antimicrobial profile (25% of marks), pharmacokinetics (25% of marks) and adverse effects (25% of marks) of Flucloxacillin and Vancomycin.
Examiner Comments
2018B 10: 49% of candidates passed this question.
Most candidates structured their answers well. Expected information included: the class of antibiotic of each agent, their respective pharmaceutics, pharmacodynamics, pharmacokinetics, indications and adverse effects. Better answers provided pharmacodynamic and pharmacokinetic information relevant to each drug rather than generic statements. Good answers also included the common resistance mechanisms for both agents.
2016A 22: 83% of candidates passed this question.
The structure required to score well was provide by the questions asked. Marks were lost by not mentioning that flucloxacillin is a beta lactam that it does not cover MRSA and that vancomycin covers enterococcus. Better answers could identify that vancomycin is slower at killing sensitive staph than flucloxacillin. Adverse effects were specifically asked for in the question so omitting facts such as associated nausea/ vomiting/ diarrhoea/ anaphylaxis etc. cost some candidates marks. If 25% of marks are allocated to side effects then it is expected more than one adverse effect would be mentioned. Some candidates had incorrect facts – Enterococcus is not a gram negative organism.
Broader
2023B 19
Describe the mechanism of action, dose, pharmacokinetics and pharmacodynamics of ceftriaxone.
Examiner Comments
2023B 19: 39% of candidates passed this question.
A structured answer under the headings of mechanism of action, dose, pharmacokinetics and pharmacodynamics worked most effectively for this question. It was expected that candidates would link the mechanism of action of Ceftriaxone (binds to PBP and inhibits final step in peptidoglycan) to its spectrum of activity. Dosing would also include indications for higher dosing, and consideration of the fact that ceftriaxone is available as an IM administration. Details on hypersensitivity (fever, nephritis, haemolytic anaemia) and consideration of C.diff infection was a main part of its pharmacodynamics. For pharmacokinetics, a structural approach is recommended, important points included excretion through both kidneys and bile and absence of liver metabolism.
2021A 16
Describe the pharmacology of piperacillin-tazobactam.
Examiner Comments
2021A 16: 62% of candidates passed this question.
Most candidates used a structured approach with the usual major pharmacology headings. Mechanism of action was well described by most, with better answers including mechanisms of resistance. Higher scoring candidates included an explanation as to the combination of the drugs. Likewise, better answers included detailed information on spectrum of activity beyond “gram positive and gram negative”, including relevant groups of organisms which are not covered. There also seemed to be some confusion about coverage for anaerobes, which piperacillin tazobactam covers well. Specific detail about adverse reactions, other than ‘allergy, rash, GI upset, phlebitis, etc’, is expected for commonly used antibiotics.
2020A 05
Outline the mechanisms of antimicrobial resistance (50% of marks).
Briefly outline the pharmacology of ciprofloxacin (50% of marks).
CICMWrecks Answer: Antibiotic resistance

MECHANISMS OF ANTIBIOTIC RESISTANCE
- Mechanisms of Antibiotic Resistance can be classified broadly:
- Efflux Pumps
- Blocked Penetration / Alteration in access to target site
- Target Modification
- Modification of Drug or pathways
- There might be multiple resistance mechanisms at play in the same organism
| Efflux Pumps: Increased efficiency or expression of efflux pumps (inner membrane proteins). | |
| Removed from cell | Active transport of ciprofloxacin out of the bacterial cell |
| E.Coli to tetracycline. | |
| Trapped between cell wall layers | glycopeptide resistance in VRSA. |
| Blocked Penetration / Alteration in access to target site | |
| Narrowing of porin channels | Streptococcal resistance to penicillins typically occurs by reducing access to PBPs. |
| Alteration of porins in gram negative outer membranes | downregulation of Outer Membrane Proteins (eg enterobacteriaceae, pseudomonas vs penems/cephs) |
| increased selectivity of Outer Membrane Proteins (eg klebsiella outbreaks) | |
| Loss of non-essential transporter channels | Anaerobes have no oxygen-transport channel which prevents penetration by aminoglycosides (Gentamicin) |
| Reduced binding of the antibiotic | VanA and VanB vancomycin resistance involves a gene mutation leading to decreased affinity of vancomycin for the binding sites of peptidoglycan precursors |
| Changes to the DNA-binding surface of DNA supergyrase infers resistance against ciprofloxacin | |
| Target Modification | |
| Modifying the enzyme that the antibiotic inhibits | Rifampicin resistance by point mutations, insertions, or deletions in RNA polymerase gene |
| Production of an alternative enzyme for that which the antibiotic inhibits | MRSA develop or acquire the gene mecA which synthesizes an additional penicillin binding protein that enables it to continue cell wall synthesis in the presence of a beta lactam drug |
| Overproduction of the target of the antibiotic | Trimethoprim DHFR enzyme overproduction in Escherichia coli and Haemophilus influenzae. |
| Synthesis of target-protecting proteins | Ribosomal protection proteins against Tetracyclines in multiple Gram positive and gram negative bacteria |
| Modification of Drug or pathways | |
| Development of metabolic pathways to bypass site of action of antibiotic | Resistance to TMP-SMX by allowing bacteria to synthesize or absorb folic acid. |
| Enzymes produced to Metabolize the drug | β-lactamases and cephalosporinases hydrolyse β-lactam rings |
| Enzymes produced that add a chemical group to antibiotic to inhibit its activity | Aminoglycoside resistance by Staphylococcus aureus or Pseudomonas (Multiple enzymes acetyltransferase, adenyltransferase, and phosphotransferase) |
Spread of Bacterial Resistance
- Selective pressure selects for favourable mutations of resistance
- mosaic genes (from other bacteria eg strep pneumo from strep mitus) or uptake of DNA from environment
- transfer of resistant bacteria from person to person
- horizontal gene transfer;
- transduction – acquisition of bacterial DNA from a phage (a virus that propagates in bacteria);
- transformation – uptake and incorporation of free DNA released into the environment by other bacterial cells;
- conjugation – gene transfer (usually on plasmids), by direct cell-to-cell contact through a bridge.
Sources:
Microbiology Lippincott Williams & Wilkins
https://courses.lumenlearning.com/microbiology/chapter/drug-resistance/
https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/
Gladwin / Sakurai / JC 2019
CICMWrecks Answer: Pharmacology: Ciprofloxacin
PHARMACOLOGY OF CIPROFLOXACIN
Examiner Comments
2020A 05: 71% of candidates passed this question.
Most candidates had a structured answer to mechanisms of resistance that covered the major categories (alter target protein, prevent entry, efflux, degrade drug) and provided an example of a bacteria and the affected antibiotic, as was required to answer the question in full. Ciprofloxacin, whilst perhaps not a first line drug in the ICU, was not well known by many candidates. Better answers included a brief outline of class, mechanism of action (action on DNA gyrase to inhibit replication), spectrum (Gram negatives particularly mentioning Pseudomonas, lesser Grampositive cover, not anaerobes, some atypical), PK (with correct dose, wide penetration into tissues including bone/prostate etc., predominantly renal excretion), side effects/toxicity (common or specific to cipro e.g. QT, tendinitis, arthropathy) and an example of resistance.
2009A 23
Describe the mechanism of action, antibacterial spectrum and pharmacokinetics of aminoglycosides.
2011A 10
Discuss the bacteriocidal activity, and toxicity, of gentamicin
CICMWrecks Answer: Aminoglycosides
AMINOGLYCOSIDES
Mechanism of Action
- Permeates into the bacterial cell down electronegative gradient (negative interior) via O2 dependent process
- Can be retarded by acidic, anaerobic environments (abscess)
- Inhibits bacterial 30S ribosomal subunit
- Inhibits bacterial protein synthesis
- Inhibition of bacterial growth (bacteriostatic) → bacteriocidal
- Synergism with cell wall inhibitors (β lactam antibiotics)
- Penicillins inhibit peptidoglycan synthesis → increased permeability to aminoglycosides → inhibition of protein synthesis (including penicillin binding protein) → Further increase in cell permeability → death
- Displays concentration dependent killing and post-antibiotic effect
- Peak plasma aminoglycoside >8 MIC required
- Large AUC not required
Antimicrobial Spectrum
- Strong activity against aerobic gram negative bacteria
- Effective against gram positives esp. with β lactam synergism
- Ineffective against obligate anaerobes
Pharmacokinetics
- Absorption
- Poor GI absorption, UNLESS inflammation or ulceration
- Distribution
- Vd approximating extracellular fluid
- Poorly protein bound
- Poor CNS, ocular penetrance
- Metabolism
- Minimal metabolism
- Elimination
- Renal clearance via filtration
- T1/2b 2~3 hours
- Can be removed by dialysis
CICMWrecks Answer: Gentamicin (Limited)
GENTAMICIN
Gentamicin
- Covers a wide range of gram negative enterobacteria and has gram positive cover that includes staph and some streptococci.
- No anaerobic activity but is synergistic with beta lactams and vancomycin.
- Used for infections of the GUT, GIT, respiratory tract, skin and soft tissues, neutropenic sepsis, CNS infections, and surgical prophylaxis
Mechanism
- Bactericidal antibiotic, which inhibits the bacterial 30S ribosomal subunit
- This impairs transcription and/or induces misreading of the mRNA, impairing protein synthesis
- diffuse across outer membranes via porins
- actively transported by oxygen dependent process across cell membrane to cytoplasm
- low O2 and extracellular pH prevent this process
Toxicity
- Gentamicin is not metabolized in humans
- It is excreted in the urine unchanged
- The presence of transport molecules in the epithelial cells of the proximal and distal tubule and the cortical collecting ducts allows gentamicin to accumulate within the cytosol of these cells.
- In the cytosol, gentamicin acts on the endoplasmic reticulum, impairing protein synthesis, and on the mitochondria to impair ATP production, increasing oxidative stress via the production of free radicals and superoxides. It also acts on lysosomes to impair protease degradation, causing further cell damage
- Independent of the cellular damage, gentamicin also inhibits some of the epithelial cell transport processes. Tubular damage may then partially or totally obstruct the lumen, causing further disruption of the nephron resorptive processes à hence the rise in plasma creatinine and fall in eGFR
- The accumulation of drug within the cell means that the toxic effects can continue long after plasma drug levels have declined
- Ototoxicity may be caused through similar processes, via accumulation of the drug in the inner ear perilymph, where it disrupts mitochondrial protein synthesis and promotes formation of free radicals within the hair cells
- Nephrotoxicity usually reverses with cessation of drug, however ototoxicity may be permanent
- Gentamicin may also cause muscle weakness due to impairment of prejunctional release of ACh, use with caution with NMDR and in myasthenia gravis.
Examiner Comments
2009A 23: Pass rate: 50%
This answer was generally well done. Most answers showed good understanding of mechanism of action. The antibacterial spectrum and pharmacokinetics were done less well.
Clinical observation guiding your study would help in answering this question, such as renal dosing, monitoring drug levels and situations in which aminoglycosides are used.
Syllabus M2 2d
Reference: Katzung 10th edition p 755-762
2011A 10:
The first part of the question on bactericidal activity of gentamicin was better answered than the second part on its toxicity. Details on the cellular mechanisms of bactericidal action and toxicity were lacking in most answers.
Most candidates did not appreciate that gentamicin is avidly accumulated and retained by proximal renal tubular cells in concentrations many times higher than the plasma concentration. Also these high tubular cell concentrations of gentamicin are maintained long after the plasma concentrations have fallen to very low levels, thus enhancing its toxic effects. Gentamicin has multiple toxic effects within the tubular cell including adverse effects on protein synthesis, translation and folding, impairment of mitochondrial function and production of reactive oxygen species and damage to the nucleus.
Syllabus: M2a, 2d. Recommended sources: Pharmacological Basis of Therapeutics, Goodman and Gillman, Chp 45 and page 1162
Mixed Comparisons
2007B 05
Compare and contrast the spectrum of activity and the mechanisms of microbial resistance for the following penicillins: benzyl penicillin, flucloxacillin and ampicillin.
Examiner Comments
2007B 05: 4 candidates (57%) passed this question.
It was expected candidates would specifically address both the spectrum of activity and mechanism of resistance.
Benzyl penicillin is highly active against Gram positive organisms, particulary streptococci but also effective against Meningococcus / Clostridia and other anaerobes, Listeria and is used in the treatment of syphilis (treponemma). It is readily hydrolysed by penicillinases or beta lactamases so any organisms that produce these are resistant i.e. most staphylococci.
Flucloxacillin contains a modified beta lactam ring so is not susceptible to hydrolysis by staphylococcal penicillinases, therefore the spectrum is Staphylococci not resistant to methicillin (i.e. not MRSA). Extra credit was given for comments that it won’t cover Listeria or some other organisms covered by Benzyl penicillin and it is much less active than Benzyl penicillin on organisms sensitive to both.
Ampicillin is an alpha amino benzyl penicillin (Aminopenicillin) and has a broader activity than Benzyl penicillin so covers the streptococci but aslo a variety of gram negative bacteria including some enterobacteriacae and Haemophilus influenzae. It also covers Helicobacter and Enterococci (better than Benzylpenicillin). It is destroyed by beta-lactamase.
Additional credit was given for discussion of other mechanisms of bacterial drug resistance.
2019B 20
Compare the pharmacology of piperacillin-tazobactam and ciprofloxacin.
Examiner Comments
2019B 20: 58% of candidates passed this question.
This question was most effectively answered using a tabular format. Only a minority of candidates demonstrated a comprehensive knowledge of these level 1 drugs and very few candidates compared the two in areas which lent themselves to comparison. The spectrum of activity generally lacked detail. Few candidates mentioned that piperacillin-tazobactam had superior gram-positive cover, both have extensive gram-negative cover including Pseudomonas.
Piperacillin-tazobactam is effective against anaerobes; whilst ciprofloxacin has some atypical cover against Mycoplasma. The mechanism of action was generally well described for piperacillin; many candidates incorrectly stated the mechanism of action for ciprofloxacin, confusing the drug with a macrolide.
Better answers included time- dependant and concentration-dependent killing. The concept of half-life was frequently confused with the dosing interval.
Minimal marks were awarded for “allergy” and “gastrointestinal side-effects”. Better candidates mentioned Liver function derangement, neutropenia, interstitial nephritis for piperacillin and tendonitis for ciprofloxacin.
2010B 03
Compare and contrast Ceftriaxone and Meropenem with respect to the following:
a. Mechanism of action and spectrum (40% of marks)
b. Pharmacokinetics (30% of marks)
c. Effect of critical illness on pharmacokinetics and subsequent dosing. (30% of marks)
Examiner Comments
2010B 03: 5 (33%) of candidates passed this question.
Most candidates used a table to compare these two drugs, which was an ideal means to structure their answers to this question and a useful way to present the information required. The use of a structured format for learning pharmacokinetic data about all drugs allows more useful information in a question such as this. Candidates should also give consideration to the effects of critical illness (such as cardiac output, plasma binding and volume of distribution) affect the pharmacokinetics of drugs used in intensive care. This particular section of the question was poorly answered.
Syllabus: M2a,2d
References: Goodman and Gillman, The Pharmacological basis of therapeutics Chp 46
2011B 22
Outline the mechanism of action of ampicillin, gentamicin, vancomycin and ciprofloxacin.
How does resistance develop to each of these antibiotics?
Examiner Comments
2011B 22: 9 (36%) of candidates passed this question.
The question could have been answered succinctly using a table. Better answers included detail of how antibiotics inhibited enzymes, effects on ribosomal subunits, and impact on cell wall synthesis or inhibit DNA gyrase. The mechanism of resistance required an understanding of beta-lactam inhibition, gene mutation altering target enzymes and ribosomal subunits, and efflux mechanisms. Mention of Van A and Van B phenotypes was also sought.
Syllabus: M2a 2c,d
Recommended sources: Rang and Dale Pharmacology Chp 46, Katzung Chp 43, 45, 46.
2013A 08
Compare and contrast the mechanism of action, spectrum of activity and adverse effects of benzyl penicillin, metronidazole and clindamycin.
Examiner Comments
2013A 08:
This question asked to compare and contrast, inviting candidates to tabulate their answers.
Details concerning other elements of pharmacology (apart from mechanism, spectrum and adverse effects) were not required and did not attract marks.
There was a lack of accurate detail in answers regarding clindamycin. Spectrum of activity mentioned by candidates was often quite narrow. The gram negative and anaerobic spectrum of activity afforded by benzyl penicillin was also not mentioned by many
candidates. Adverse reactions were an opportunity to score marks – all drugs can cause
nausea, vomiting, rash and hypersensitivity phenomena – especially the antibiotics. However it is important that specific side effects for each agent are also mentioned by candidates.
Information that related to the pharmaceutics, pharmacokinetic or pharmacodynamic
properties of these drugs was not requested and did not score marks.
ii. Describe the classification and pharmacology of antiviral and antifungal agents.
2010A 12
Outline the classification of viruses giving examples of each class (60% marks). Describe the mechanism of action of acyclovir and oseltamivir (40% marks).
CICMWrecks Answer
Baltimore Classification system (for Viruses)

| I | dsDNA viruses | mRNA is transcribed directly from the DNA template | Adenoviruses, Herpesviruses, Poxviruses |
| II | ssDNA viruses | (+ strand or “sense”) DNA DNA is converted to double stranded form before RNA is transcribed | Parvovirus (B19) |
| III | dsRNA viruses | mRNA is transcribed from the RNA genome | Rotavirus |
| IV | (+)ssRNA viruses | (+ strand or sense) RNA Genome functions as mRNA | Picornaviruses (Entero, HepA), Flaviviruses (Dengue) |
| V | (−)ssRNA viruses | (− strand or antisense) RNA mRNA is transcribed from the RNA genome | Orthomyxoviruses (Influenza) |
| VI | ssRNA-RT viruses | (+ strand or sense) RNA with DNA intermediate in life-cycle RT makes DNA from the RNA genome DNA is then incorporated into host genome mRNA is transcribed from the incorporated DNA | Retroviruses (HIV) |
| VII | dsDNA-RT viruses | DNA with RNA intermediate in life-cycle Viral DNA is replicated through an RNA intermediate, the RNA may serve directly as mRNA or as a template to make mRNA | Hep B |
Key:
ss: Single stranded
ds: Double stranded
(-): – strand or antisense
(+): + strand or sense
RT – Reverse Transcriptase
Past Classifications (now not used):
By shape:
- filamentous
- isometric (or icosahedral)
- envelopedH
- Head and tail
By Genome Structure and Core:
- RNA / DNA
- Single-stranded / Double stranded
- Linear/Circular
- Non-segmented/Segmented
By capsid structure:
- Naked icosahedral – Hep A, Polio
- Enveloped icosahedral – EBV, HSV
- Enveloped helical – Influenza, Measles
- Naked Helical – Tobacco Mosaic virus
- Complex – Herper, Smallpox, Hep B
Mechanism of Action: Oseltamivir vs Aciclovir
| Oseltamivir | Aciclovir |
|---|---|
| Neuraminidase inhibitor (Anti-Influenza) | Viral guanosine analogue |
| Prodrug | Prodrug |
| Active metabolite inhibits viral Neuraminidase | Converted to aciclovir monophosphate by viral thymidine kinase which in turn is converted to aciclovir triphosphate by host cell |
| cleaves the sialic acid (which is found on glycoproteins on the surface of human cells that helps new virions to exit the cell) | ACV-TP is HSV-specified DNA polymerase inhibitor |
| prevents viral release by infected cells (viral shedding) | inhibits nucleic acid production, incorporates into DNA resulting in chain termination |
| Active against Influenza | Active against herpesvirus family: HSV-1,2, VZV,EBV,CMV(least) |
Mooney / JC 2020
Examiner Comments
2010A 12: 1 (10%) of candidates passed this question
Viruses are classified according to:
a) genetic material
b) mode replication
c) structural proteins (capsids)
d) presence of an envelope
Thus DNA viruses, double or single stranded DNA usually replicate in the nucleus of the host cell via polymerase, not incorporated into the host genetic material. Examples being double stranded, herpes, adenovirus, poxvirus and single stranded, parvovirus. In comparison, RNA viruses, single strand and have 2 different reproduction strategies, being RNA sense(positive) and RNA antisense(negative), an example is paramyxovirus. For retroviruses, the single stranded RNA can’t act as mRNA and is transcribed into DNA by a reverse transcriptase. This DNA is incorporated into the host DNA, so the host makes the viral RNA, for example HIV.
Candidates were also expected to briefly mention capsids and viral envelopes. In relation to the second part of the question, candidates were expected to mention that acyclovir inhibits DNA polymerase in the terminal nucleic acid chain and that oseltamivir is a neuraminidase inhibitor which prevents the budding of new viruses from the infected cells. Most candidates had very little knowledge of this area.
Syllabus: M2 2a&d 8 Reference: Medical Microbiology and Infection at a Glance, Gillespie &, Bamford pgs 58,59, Basic and Clinical Pharmacology, Katzung pgs791,815
2021B 04
Compare the pharmacology of fluconazole and amphotericin.
Examiner Comments
2021B 04: 6% of candidates passed this question.
This question exposed an area of the syllabus neglected by the candidates. Answers were generally vague in detail with lots of incorrect facts and generally displayed a very limited knowledge. Antifungal agents are regularly used in critically ill patients either as treatment or prophylaxis. An understanding of the aspects of these drugs with respect to spectrum of activity, mechanism of action, specific PK and PD properties as well as potential side effects would have been the basis for this compare and contrast question. Examiners want to be convinced that the candidates understand the strengths and weaknesses of each drug and in which circumstances one agent might be used in preference to the other.
2016B 23
Compare and contrast the mechanism of action, spectrum of activity and adverse effects of benzyl penicillin and fluconazole.
Examiner Comments
2016B 23: 8% of candidates passed this question.
To pass this question each of the three components needed to be compared and contrasted for both agents. A tabulated answer helped in this regard but was not essential.
Some answers included information that could not gain marks, as it was not directly relevant to the question asked (e.g. presentation and dose).
In spectrum of activity, as well as what important organisms the agents were effective against, marks were also given for the important organisms that they were not effective against (e.g. MRSA and beta-lactamase producing organisms for penicillin G; and aspergillus for fluconazole).
In general, of the two agents, fluconazole was the least well answered. For example, a common omission either in mechanism of action or in adverse effects was that fluconazole inhibits microsomal P450 enzymes.
Some candidates confused fluoroquinolone with fluconazole.
iii. Outline the pharmacology of antiseptics and disinfectants.
2008A 14
Define the terms antiseptic and disinfectant. Briefly describe the advantages and disadvantages of alcohol, chlorhexidine, glutaraldehyde and povidone iodine.
CICMWrecks Answer
- Disinfectants
- Strong chemical compounds that kill of inhibit the growth of microbes
- Antiseptic
- Disinfectants with sufficiently low toxicity to host cells, that can be used directly on skin, wounds or mucosa
Pharmacopeia - Antiseptics & Disinfectants (Level 3)
PC: Pharmaceutics, PD: Pharmacodynamics, PK: PharmacokineticsColumn Visibility: To filter out some of the drugs
Dropdown Menus: To filter out categories or items
Search bar: To search within table
Print: Print to PDF or printer. Pls remember to change to landscape if the table does not fit in portrait mode.
| CATEGORY | ITEM | ALCOHOL | CHLORHEXIDINE | IODINE | GLUTARALDEHYDE |
|---|---|---|---|---|---|
| Basics | CICM Level of Understanding | Level 3 | Level 3 | Level 3 | - |
| PD | Main Action | Likely act by denaturing proteins | Cationic biguanide -strongly adsorbs to bacterial membranes causing leakage of small particles and precipitation of cytoplasmic proteins | Oxidative damage. | Alkylation of micro-organism proteins or nucleic acids |
| PD | Mode of Action | Evaporative effects are useful when sinks with running water are not available | Resistant to inactivation by blood and organic material Low skin irritation | 1. Sporicidal 2. Cheap 3. Broad spectrum Most effective for intact skin | Used for sterilization of equipment that cannot withstand high temperatures |
| PD | Route & Doses | 1. Flammable- must be allowed to dry fully before diathermy or laser surgery 2. Corneal damage 3. Skin drying 4. Ineffective against C. Dif spores | 1. Neurotoxic 2. Delayed effect 3. No direct spore activity | 1. Hypersensitivity reactions 2. Delayed onset without residual activity 3. Stains clothes and dressings | 1. Once activated have a shelf life of 14 days 2. Failure of activity may occur due to excessive dilution or exposure to organic material (frequently reused) 3. Irritating to eyes and respiratory mucosa 4. Slow activity |
| PD | Spectrum of Activity | Onset: Rapid Duration: Lack residual action because they evaporate completely | Onset: Delayed Duration: Sustained residual activity | Onset: Iodine is bacteriocidal in 1 minute and kills spores in 15 minutes. However in povidine compounding it has a delayed onset Duration: No sustained effect | Onset: 2.4% glutaraldehyde achieve disinfection in 45 minutes |
| PD | Resistance Mechanism | - Gram positives and negatives - Acid fast bacteria are susceptible - Lipophilic viruses may be susceptible - Many fungi | - Bacteria (G+ve>G-ve) - Moderate fungal and viral activity - Inhibits spore germination | - Bacteria (G+ve and –ve and acid fast) - Sporicidal - Viruses - Fungi | - Bacteria (G+ve and –ve and acid fast) - Spores - Viruses - Fungi |
| PD | Ineffective Against: | Spores and prions Hydrophilic viruses are less susceptible | Spores and prions Hydrophilic viruses are less susceptible | Prions Hydrophilic viruses | Prions |
| Spl | SPECIAL POINTS | Optimal bacteriocidal concentration is 60-90% | Can be combined with 70% alcohol Preferred antiseptic for central venous access Not absorbed orally | Used to sterilize fiber optic endoscopes and respiratory therapy equipment |
Sakurai 2016
Examiner Comments
2008A 14: No candidates (0%) passed this question.
It was expected candidates could define and distinguish between these terms with a specific comment that disinfectants are applied to inanimate objects and antiseptics can be applied to living tissue. The advantages and disadvantages could be addressed either tabulated or discussed in point form, either was acceptable.
It was expected answers would include a comment on each agent and specifically address areas such as general spectrum of activity, speed of onset ( agents that need to dry to be effective versus those with more rapid onset), duration of effect ( residual activity), limitations of use and potential hazards. Marks were awarded for identifying Glutaraldehyde as a disinfectant (as opposed to the other antiseptic agents) and its use for cleaning equipment such as endoscopes with the precautions required for potential toxicity.
Additional credit was given for discussion of relevant facts such as the proven benefit for chlorhexidine skin preparation for central venous line insertion. Candidates are referred to several of the recommended texts which cover this area well.
VIVAs
| 2023B | MoA of antibiotics with examples |
| 2023A | Factors that determine ability of IV Antibacterial to treat localized infection |
| 2022B | |
| 2022A | Pharmacological properties to consider when chosing alternative option to meropenem with examples Gentamicin pharmacology |
| 2021B | Bacteria gram stain, antibiotics |
| 2021A | |
| 2020B | Aminoglycoside pharmacology, Gentamicin MoA Bacteria gram stain, antibiotics |
| 2019B | |
| 2019A | |
| 2018B | Microbial pathogen classification |
| 2018A | |
| 2017B | classification of micro-organisms and antimicrobial pharmacology. What microorganisms are demonstrated on Gram stain |
| 2017A | classification of bacteria. |
| 2016B | |
| 2016A | Antibacterial agents, resistance, dosing rationale for aminoglycosides |
| 2015B | Classicication and mechanisms of Abx resistance. Immunology related to anaphylaxis and immunoglobulins |
| 2015A | |
| 2014B | |
| 2014A | |
| 2013B | |
| 2013A | |
| 2012B | |
| 2012A | Classify bacteria, antibiotics, gentamicin |
| 2011B | |
| 2011A | |
| 2010B | |
| 2010A | |
| 2009B | |
| 2009A | How do bacteria differ from the majority of normal human cells (eukaryotes)? Antimicrobial agents. |
| 2008B | |
| 2008A | How are bacteria classified? |
| 2007B |

Recent Comments